SimpleClade
By Dave Dobson
(ddobson@guilford.edu)

SimpleClade is a software program that allows simple cladistic analysis with a graphical user interface. I designed it for use in my paleontology class to let students play around in a straightforward way with cladistics. It is designed to be instructive and easy to use, not to be a research tool. It is still under development, but I've used it successfully in an introductory lab. It is available free of charge, but please let me know by e-mail at ddobson@guilford.edu if you try it out and if you have any suggestions.
Click Here to Download SimpleClade (217 kb zipped archive)
Brief User Guide
Installation: Extract the program and associated files from the compressed ZIP archive.
Data Files: Create your own data file, or use one of the three provided. The data files must be in CSV (comma-separated value) format, which is easily done in Excel and other software programs. The file DATA.XLS included with the program has a template for data entry. Make sure to leave unused cells marked "UNUSED" as shown in those files and in the images below. The program currently supports up to 20 species (or any organism classification type) and 20 traits. If you don't use all 20 species or traits, the data you enter must be in the left-most and uppermost rows, and there must be at least one "UNUSED" tag to the right of your data and below your data. The data values can be any text or numeric value, but if you use numbers, the numbers will not be used in any ordinal or scalar sense - each number will just be a unique trait state (i.e. the difference between 1 and 3 is the same as the difference between 7 and 1000 and the difference between hairy and smooth).
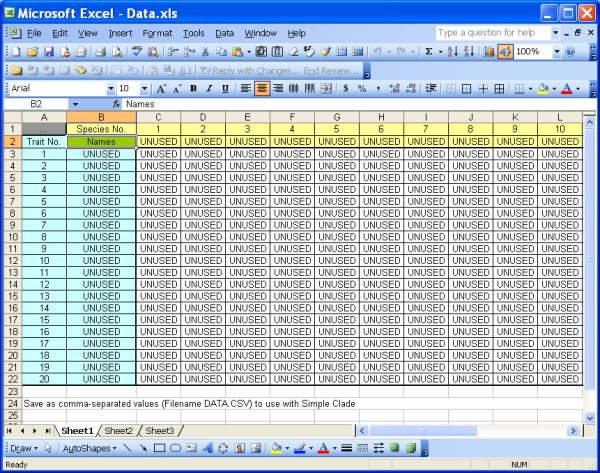
Data.xls file - template for data editing
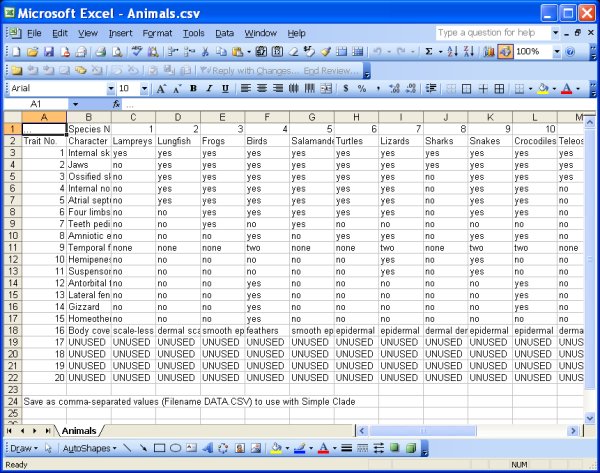
Animals.csv file - example of file saved in CSV format
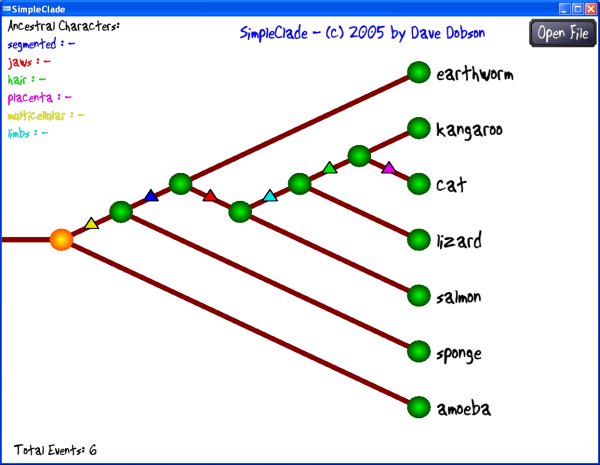
Program Operation: Click on the "Open File" button to open your data file. If your file is formatted properly, SimpleClade should read it in and display the information as a cladistic tree diagram. Initially, the relationships specified will be in the order you list them in your file.
Selecting a node: Click on any green node circle to select it. The trait states associated with that particular node will be displayed.
Changing a connection: Once you've selected a node, you can disconnect it and reconnect it to any branch on your tree that is a legal reconnection. The connections you can make are shown as curved lines with a branching symbol on the limb to which you are connecting. Click the mouse to move your selected node to the indicated location.
Parsimony: SimpleClade calculates a simple parsimony statistic - the number of trait state changes in your tree. This is listed as Total Events in the lower left corner of the window. The goal of a cladistic analysis is generally to minimize the total trait state changes, thereby establishing the simplest (though not necessarily the correct) phylogenetic pathway for the traits and genera under study.
Outgroup: To make a particular organism your outgroup, connect it to the horizontal root stem of the tree on the left side of the screen. The traits of that organism will be listed as in the Ancestral Traits section at the left side of the screen.
Viewing trait state changes: Each trait state change is marked with a triangle (delta) on the branch of the tree where it changes. If you move your mouse cursor over a triangle, the trait state change will be listed at the lower right on the screen.
Missing Features
It is not possible at this point to save or print your work directly - the program was designed merely to be used as a class exercise. I may add these features if there is interest in them. However, it is easy to get a screen capture of your cladistic tree - just hold down alt and hit the PrintScreen key, and the window image will be copied to memory. You can then paste it into a word processing or image editing program and store or print it that way.
It is not possible to weight certain traits more than others, to have some trait states be counted as more similar than others, or to do any of the other complex routines often associated with cladistic analysis. If you want to see such a feature, then let me know and I'll see how hard it would be to add it.
Please let me know if you try it out and what you think. If you create a data file that you'd like to share, please send it to me (ddobson@guilford.edu) so I can post it here - the more examples I can send out with the program, the more useful it will be to students.